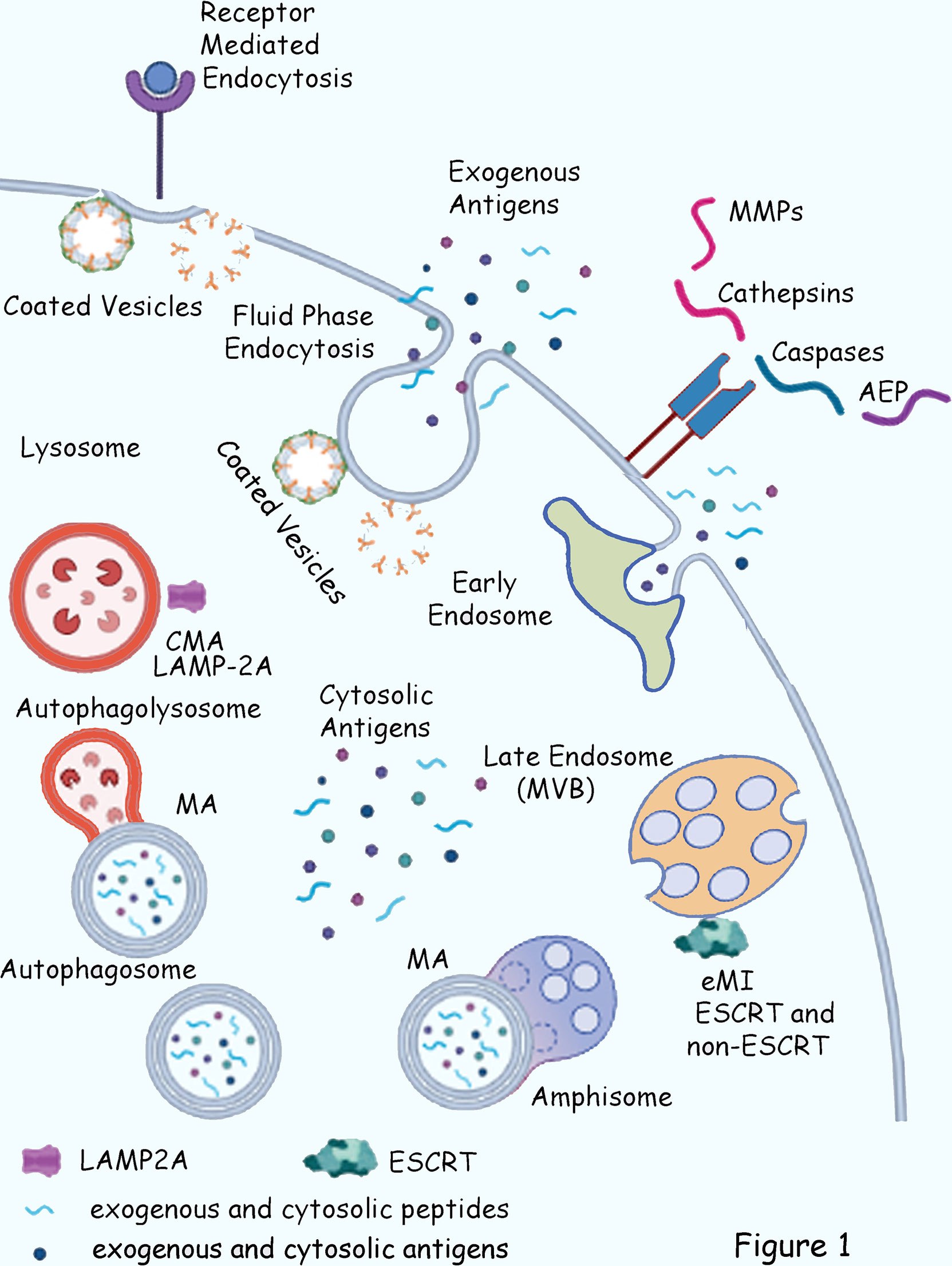
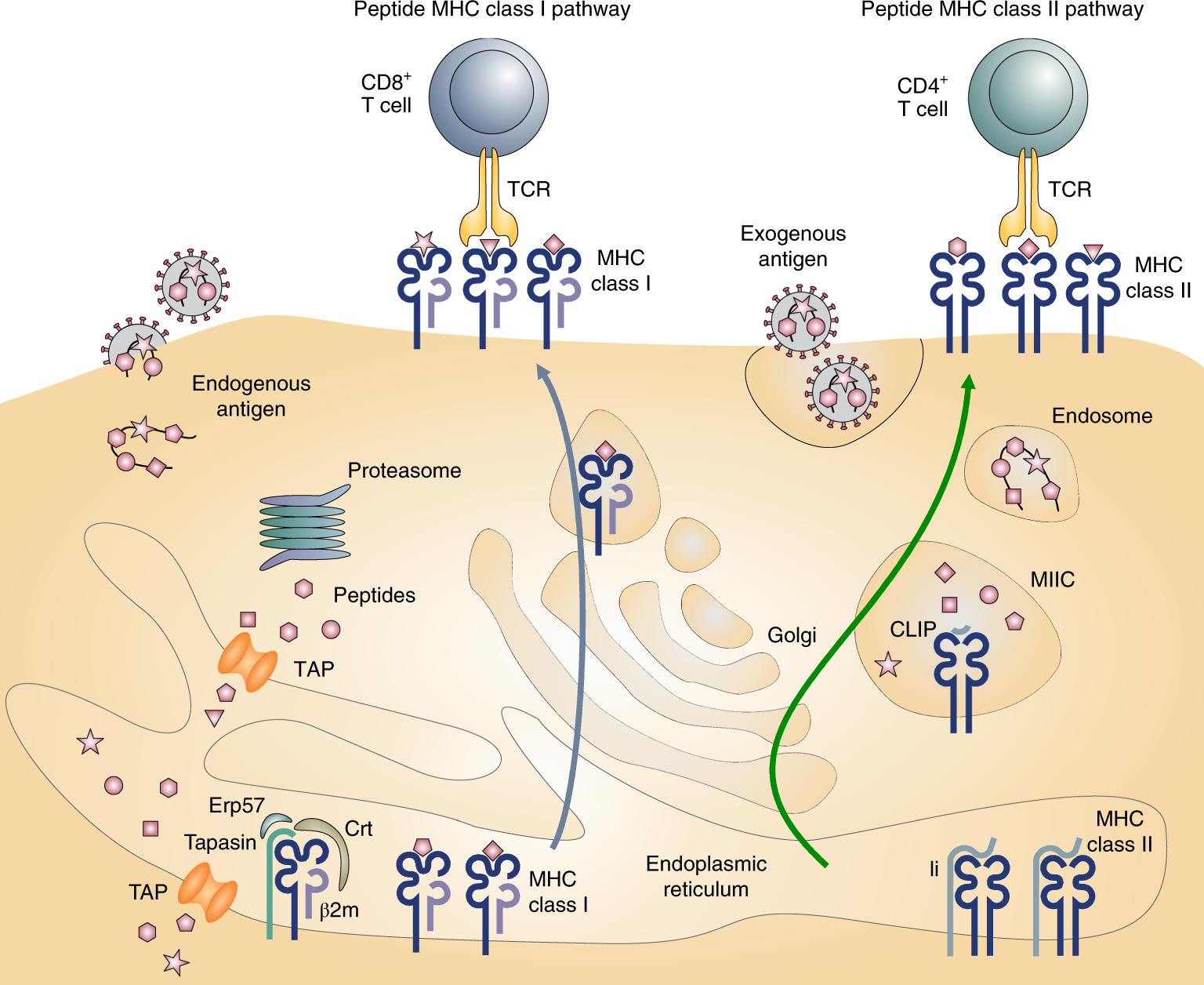
Endosomal processing of antigens and major histocompatibility complex... | Download Scientific Diagram

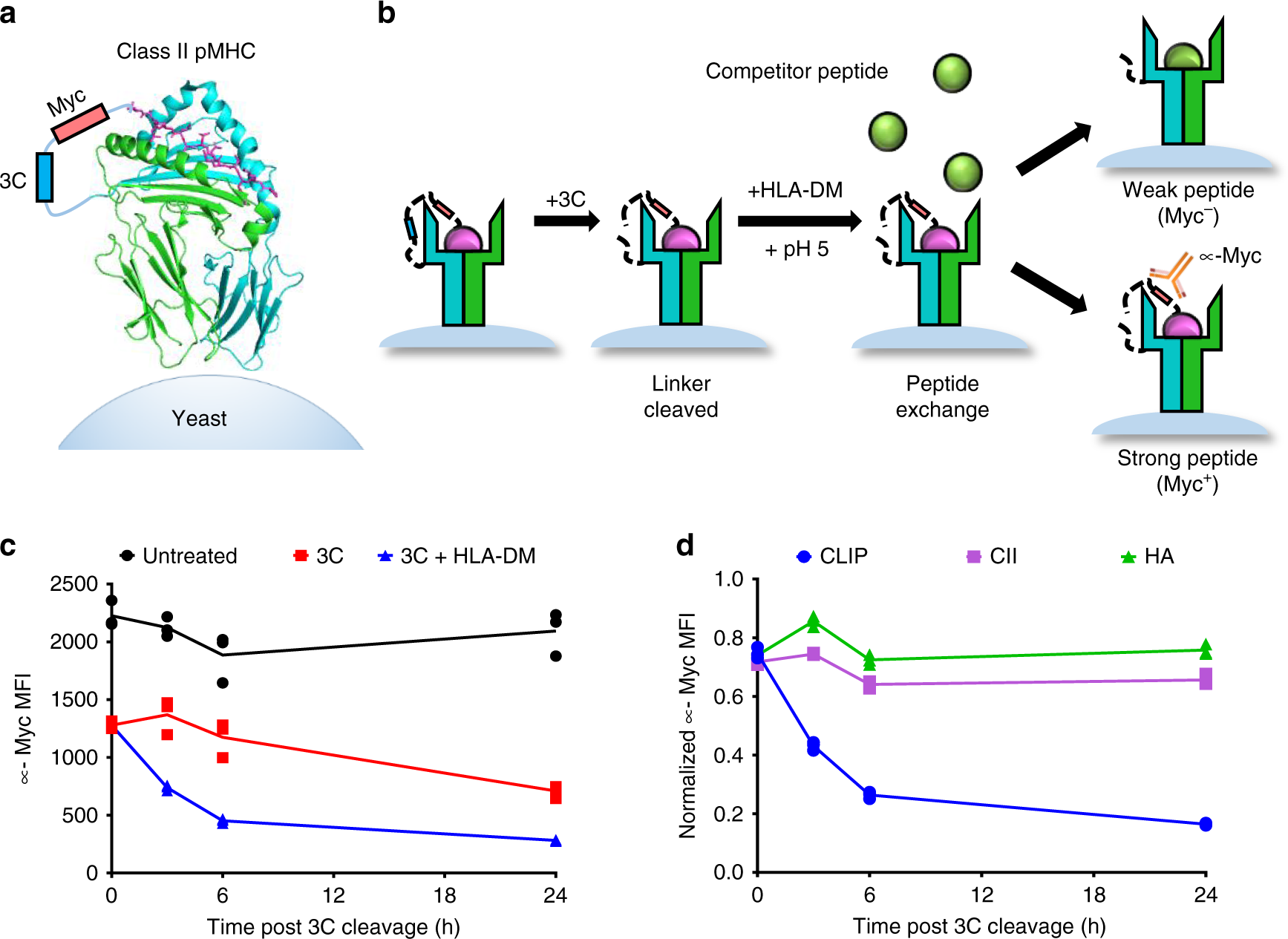
The mechanism of HLA-DM induced peptide exchange in the MHC class II antigen presentation pathway. | Semantic Scholar
Mass spectrometry–based identification of MHC-bound peptides for immunopeptidomics | Nature Protocols
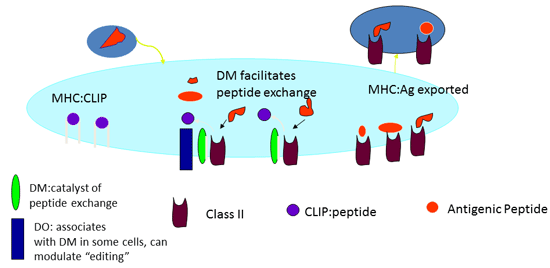
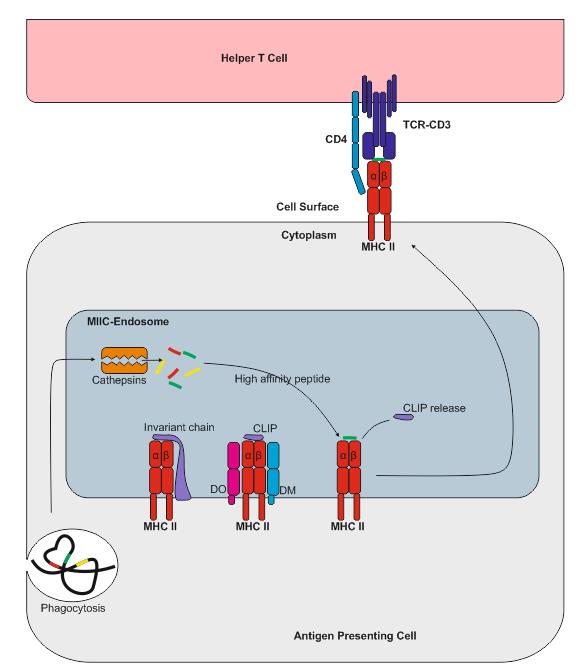
The Role of DM in Selecting Epitopes for CD4 T Cell Responses - Projects - Sant Lab - University of Rochester Medical Center

Deciphering the Structural Enigma of HLA Class-II Binding Peptides for Enhanced Immunoinformatics-based Prediction of Vaccine Epitopes | Journal of Proteome Research

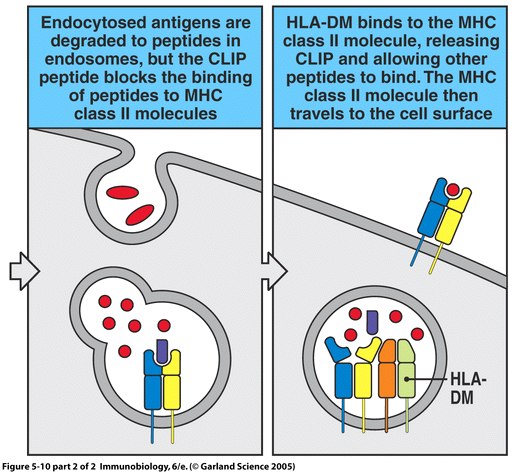
Endosomal compartment: Also a dock for MHC class I peptide loading - Ma - 2014 - European Journal of Immunology - Wiley Online Library